R tidyverse: dplyr.

dplyr
dplyr is a package from tidyverse and its purpose is sorting and configuring your data as you please and prepare the data before analysis.
In this tutorial we will need first load the tidyverse package,(there is no need to load dplyr it is one of the tidyverse cores) and then we will load nycfights13 package just to get the dataset flights to work with this.
# Install packages if it is your firts time.
install.packages("tidyverse")
install.packages("nycfights13")
# load packages
library(tidyverse)
library(nycflights13)
Filter rows with filter()
filter() function allows you to subset a data if some condition is fulfilled.
Example subsetting the day 01 from dataset flights, but first we will see how is our data:
dim(flights)
## 336776 19
dim() function show us that our data has 336.776 rows and 19 columns.
Now we are going to use filter() function.
Day_01 <- filter(flights, month ==1 & day == 1 )
dim(Day_01)
## [1] 842 19
You can add an “OR” condition using “|”, in th next example we are going to filter all flights in January OR February
Jan_Feb <- filter(flights, month == 1 | month == 2)
#or use %in% to set a shortcut
Jan_Feb <- filter(flights, month %in% c(1,2))
Also, you can add an AND condition with “&”, we are going to filter all flights made in January and by the plane with tailnumber “N14228”.
Jan_by_N14228 <- filter(flights, month == 1 & tailnum == "N14228")
dim(Jan_by_N14228)
## [1] 15 19
There were just 15 flights made in January by the flight “N14228”.
Exclude data
If you need to exclude part of your data when a condition is fulfilled you should use !.
without_Jan <- filter(flights, !(month == 1))
Arrange Rows with arrange()
To arrange data in your dataset you must indicate the column to arrange and dplyr will do it.
dataset_sorted <- arrange(flights, month, day, year)
Reordering by a column in descending order
| year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2013 | 1 | 1 | 517 | 515 | 2 | 830 | 819 | 11 | UA | 1545 | N14228 | EWR | IAH | 227 | 1.4e+03 | 5 | 15 | 2013-01-01 05:00:00 |
| 2013 | 1 | 1 | 533 | 529 | 4 | 850 | 830 | 20 | UA | 1714 | N24211 | LGA | IAH | 227 | 1.42e+03 | 5 | 29 | 2013-01-01 05:00:00 |
| 2013 | 1 | 1 | 542 | 540 | 2 | 923 | 850 | 33 | AA | 1141 | N619AA | JFK | MIA | 160 | 1.09e+03 | 5 | 40 | 2013-01-01 05:00:00 |
| 2013 | 1 | 1 | 544 | 545 | -1 | 1004 | 1022 | -18 | B6 | 725 | N804JB | JFK | BQN | 183 | 1.58e+03 | 5 | 45 | 2013-01-01 05:00:00 |
| 2013 | 1 | 1 | 554 | 600 | -6 | 812 | 837 | -25 | DL | 461 | N668DN | LGA | ATL | 116 | 762 | 6 | 0 | 2013-01-01 06:00:00 |
descending <- arrange(flights, desc(year))
head(descending, 5) #head() function displys just the firts 5 rows
Selecting columns with select()
This function is especially useful to subset dataframes selecting only the specified columns.
subset_columns <- select(flights, year, month,)
head(subset_columns,5)
| year | month |
|---|---|
| 2013 | 1 |
| 2013 | 1 |
| 2013 | 1 |
| 2013 | 1 |
| 2013 | 1 |
Also you can select a range within the dataset and extract it.
subset_range <- select(flights, month:dep_delay)
head(subset_range)
| month | day | dep_time | sched_dep_time | dep_delay |
|---|---|---|---|---|
| 1 | 1 | 517 | 515 | 2 |
| 1 | 1 | 533 | 529 | 4 |
| 1 | 1 | 542 | 540 | 2 |
| 1 | 1 | 544 | 545 | -1 |
| 1 | 1 | 554 | 600 | -6 |
| 1 | 1 | 554 | 558 | -4 |
Or exclude some colums with -
subset_excluded <- select(flights, -(month:distance))
head(subset_excluded)
| year | hour | minute | time_hour |
|---|---|---|---|
| 2013 | 5 | 15 | 2013-01-01 05:00:00 |
| 2013 | 5 | 29 | 2013-01-01 05:00:00 |
| 2013 | 5 | 40 | 2013-01-01 05:00:00 |
| 2013 | 5 | 45 | 2013-01-01 05:00:00 |
| 2013 | 6 | 0 | 2013-01-01 06:00:00 |
| 2013 | 5 | 58 | 2013-01-01 05:00:00 |
Improve the selection
There are some other functions that can be used with select() to improve the results, like start_with(), ends_with() and contains(), for example:
subset_by_start <- select(flights, starts_with("dep"))
head(subset_by_start)
| dep_time | dep_delay |
|---|---|
| 517 | 2 |
| 533 | 4 |
| 542 | 2 |
| 544 | -1 |
| 554 | -6 |
| 554 | -4 |
If you want to change the column order in a dataframe, you can do it adding the function everything() clarifying which columns you want first.
subset_firts_day.month <- select(flights, day, month, everything())
head(subset_firts_day.month)
| day | month | year | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 2013 | 517 | 515 | 2 | 830 | 819 | 11 | UA | 1545 | N14228 | EWR | IAH | 227 | 1.4e+03 | 5 | 15 | 2013-01-01 05:00:00 |
| 1 | 1 | 2013 | 533 | 529 | 4 | 850 | 830 | 20 | UA | 1714 | N24211 | LGA | IAH | 227 | 1.42e+03 | 5 | 29 | 2013-01-01 05:00:00 |
| 1 | 1 | 2013 | 542 | 540 | 2 | 923 | 850 | 33 | AA | 1141 | N619AA | JFK | MIA | 160 | 1.09e+03 | 5 | 40 | 2013-01-01 05:00:00 |
| 1 | 1 | 2013 | 544 | 545 | -1 | 1004 | 1022 | -18 | B6 | 725 | N804JB | JFK | BQN | 183 | 1.58e+03 | 5 | 45 | 2013-01-01 05:00:00 |
| 1 | 1 | 2013 | 554 | 600 | -6 | 812 | 837 | -25 | DL | 461 | N668DN | LGA | ATL | 116 | 762 | 6 | 0 | 2013-01-01 06:00:00 |
| 1 | 1 | 2013 | 554 | 558 | -4 | 740 | 728 | 12 | UA | 1696 | N39463 | EWR | ORD | 150 | 719 | 5 | 58 | 2013-01-01 05:00:00 |
Add new columns with mutate()
There are moments when you need add a new variable in a dataframe, and this variable is the result of other variables in the original dataframe, namely, you can create a new column from others without delete the originals.
mutated_dataset <- mutate(flights, gain = arr_delay - dep_delay)
mutated_variable <- select(mutated_dataset, gain, everything())
head(mutated_dataset) #see by yourself
Replace values with mutate()
head(iris)
my_iris <- iris %>% mutate(Species = str_replace(Species, "setosa", "SETOSA"))
head(my_iris)
| Sepal.Length | Sepal.Width | Petal.Length | Petal.Width | Species |
|---|---|---|---|---|
| 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 4.9 | 3 | 1.4 | 0.2 | setosa |
| 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 5 | 3.6 | 1.4 | 0.2 | setosa |
| 5.4 | 3.9 | 1.7 | 0.4 | setosa |
| Sepal.Length | Sepal.Width | Petal.Length | Petal.Width | Species |
|---|---|---|---|---|
| 5.1 | 3.5 | 1.4 | 0.2 | SETOSA |
| 4.9 | 3 | 1.4 | 0.2 | SETOSA |
| 4.7 | 3.2 | 1.3 | 0.2 | SETOSA |
| 4.6 | 3.1 | 1.5 | 0.2 | SETOSA |
| 5 | 3.6 | 1.4 | 0.2 | SETOSA |
| 5.4 | 3.9 | 1.7 | 0.4 | SETOSA |
Add new colums with transmutate
When you want create new columns but delete the originals, then the best options is use trasnmutate() function.
subset_transmutate <- transmute(flights, speed = (distance / air_time) / 60, year, month)
head(subset_transmutate)
| speed | year | month |
|---|---|---|
| 0.103 | 2013 | 1 |
| 0.104 | 2013 | 1 |
| 0.113 | 2013 | 1 |
| 0.144 | 2013 | 1 |
| 0.109 | 2013 | 1 |
| 0.0799 | 2013 | 1 |
Note that all the original variables have been eliminated and only remain which were enunciated explicitly.
Grouping summaries with sumarize()
This function can collapses a whole data frame in a row and it is very useful to extract summaries, for example you can calculate what is the busiest day throughout all the flies.
busiest_day <- summarize(flights, busiest_day = median(day, na.rm = T))
busiest_day
| busiest_day |
|---|
| 16 |
Grouping with group_by()
If you want extract a summary for some categories in a variable then the function group_by() is what you need.
summary_by_month <- group_by(flights, month)
summarize(summary_by_month, mean_air_time = mean(air_time, na.rm = T)) %>% transmute(month.name, mean_air_time)
| month.name | mean_air_time |
|---|---|
| January | 154 |
| February | 151 |
| March | 149 |
| April | 153 |
| May | 146 |
| June | 150 |
| July | 147 |
| August | 148 |
| September | 143 |
| October | 149 |
| November | 155 |
| December | 163 |
Combining multiple operation with pipe
There are different ways to write code in but you should avoid redundancy as much as you can to make your code more readable, one step on this route is use pipes (%>%), they can bind multiple functions and then process your object.
This is some code without %>%
by_Specie <- group_by(iris, Species)
counts <- summarize(by_Specie,
count = n())
counts_arranged <- arrange(counts, desc(Species))
The same expression can be done using %>%
counts_arranged <- iris %>%
group_by(., Species) %>%
summarize(.,count = n()) %>%
arrange(.,desc(Species))
#the dots are optional, they just represent the last object processed
Dealing with missin values
Any math operation against with a NA will generate another NA, they are infectious and tends to propagate themselves, so is useful delete them with na.rm argument when we are preparing the data.
mean_dep_delay_by_day <- flights %>% group_by(month, year, day) %>% summarize(mean = mean(dep_delay))
head(mean_dep_delay_by_day)
| month | year | day | mean |
|---|---|---|---|
| 1 | 2013 | 1 | |
| 1 | 2013 | 2 | |
| 1 | 2013 | 3 | |
| 1 | 2013 | 4 | |
| 1 | 2013 | 5 | |
| 1 | 2013 | 6 |
Solution:
mean_dep_delay_by_day <- flights %>% group_by(month, year, day) %>% summarize(mean = mean(dep_delay, na.rm = T))
Counts
As part of the example, first we will filter some data, and then create a frequency polygon removing NAs
not_cancelled <- flights %>%
filter(!is.na(dep_delay), !is.na(arr_delay))
delays <- not_cancelled %>%
group_by(tailnum) %>%
summarize(delay = mean(arr_delay, na.rm = T), n= n())
ggplot(delays, aes(n, delay))+ geom_point(alpha = 1/10)
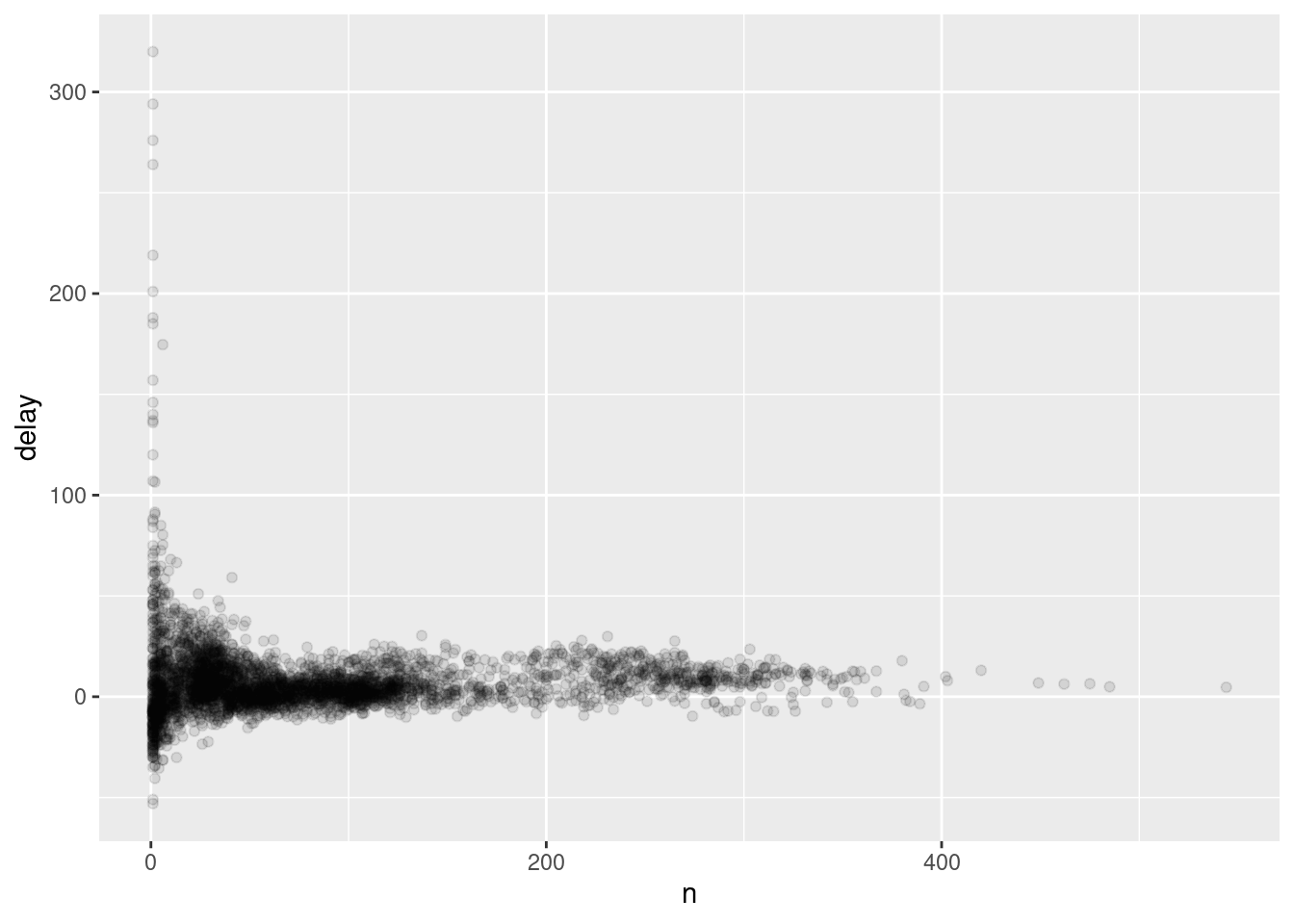
# Graphic made including counts with n()
n() function is very useful but it can not take arguments, just count a variable associated with another function, another option is use count() or table().
flights %>% count(month)
# counting with table, the information on the output is the same.
table(flights$month)
| month | n |
|---|---|
| 1 | 27004 |
| 2 | 24951 |
| 3 | 28834 |
| 4 | 28330 |
| 5 | 28796 |
| 6 | 28243 |
| 7 | 29425 |
| 8 | 29327 |
| 9 | 27574 |
| 10 | 28889 |
| 11 | 27268 |
| 12 | 28135 |
| Var1 | Freq |
|---|---|
| 1 | 27004 |
| 2 | 24951 |
| 3 | 28834 |
| 4 | 28330 |
| 5 | 28796 |
| 6 | 28243 |
| 7 | 29425 |
| 8 | 29327 |
| 9 | 27574 |
| 10 | 28889 |
| 11 | 27268 |
| 12 | 28135 |
Relational data with dplyr
Mutating joints
This tools allows you o combine two tables that have observations in common,in the next tibble you can see the variable carrier, on the other hand the tibble airlines have the same variable, then we can use the left_joint() to unite them
flights2 <- flights %>%
select(year:day, hour, origin, dest, tailnum, carrier)
head(flights2)
| year | month | day | hour | origin | dest | tailnum | carrier |
|---|---|---|---|---|---|---|---|
| 2013 | 1 | 1 | 5 | EWR | IAH | N14228 | UA |
| 2013 | 1 | 1 | 5 | LGA | IAH | N24211 | UA |
| 2013 | 1 | 1 | 5 | JFK | MIA | N619AA | AA |
| 2013 | 1 | 1 | 5 | JFK | BQN | N804JB | B6 |
| 2013 | 1 | 1 | 6 | LGA | ATL | N668DN | DL |
| 2013 | 1 | 1 | 5 | EWR | ORD | N39463 | UA |
airlines
flights2 %>%
select(-origin, -dest, - day) %>%
left_join(airlines, by = "carrier") %>% select(carrier, name , everything()) %>% head()
airlines
flights2 %>%
select(-origin, -dest, - day) %>%
left_join(airlines, by = "carrier") %>% select(carrier, name , everything()) %>% head()
| carrier | name |
|---|---|
| 9E | Endeavor Air Inc. |
| AA | American Airlines Inc. |
| AS | Alaska Airlines Inc. |
| B6 | JetBlue Airways |
| DL | Delta Air Lines Inc. |
| EV | ExpressJet Airlines Inc. |
| F9 | Frontier Airlines Inc. |
| FL | AirTran Airways Corporation |
| HA | Hawaiian Airlines Inc. |
| MQ | Envoy Air |
| OO | SkyWest Airlines Inc. |
| UA | United Air Lines Inc. |
| US | US Airways Inc. |
| VX | Virgin America |
| WN | Southwest Airlines Co. |
| YV | Mesa Airlines Inc. |
| carrier | name | year | month | hour | tailnum |
|---|---|---|---|---|---|
| UA | United Air Lines Inc. | 2013 | 1 | 5 | N14228 |
| UA | United Air Lines Inc. | 2013 | 1 | 5 | N24211 |
| AA | American Airlines Inc. | 2013 | 1 | 5 | N619AA |
| B6 | JetBlue Airways | 2013 | 1 | 5 | N804JB |
| DL | Delta Air Lines Inc. | 2013 | 1 | 6 | N668DN |
| UA | United Air Lines Inc. | 2013 | 1 | 5 | N39463 |
Now, we have added the information from the tibble airlines in a new tibble only.
There are some type of joints like inner joints (exclude the mismatches using the inner_joint() function) and outer joints who conserved the mismatched fulling the gaps with NAs.
- A left join keeps all observations in x. left_joint_()
- A right join keeps all observations in y. right_joint_()
- A full join keeps all observations in x and y. full_joint_()
Defining keys
If you do not use the argument by then by default dplyr will search for columns in common.
flights2 %>%
left_join(weather) %>% head()
| year | month | day | hour | origin | dest | tailnum | carrier | temp | dewp | humid | wind_dir | wind_speed | wind_gust | precip | pressure | visib | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2013 | 1 | 1 | 5 | EWR | IAH | N14228 | UA | 39 | 28 | 64.4 | 260 | 12.7 | 0 | 1.01e+03 | 10 | 2013-01-01 05:00:00 | |
| 2013 | 1 | 1 | 5 | LGA | IAH | N24211 | UA | 39.9 | 25 | 54.8 | 250 | 15 | 21.9 | 0 | 1.01e+03 | 10 | 2013-01-01 05:00:00 |
| 2013 | 1 | 1 | 5 | JFK | MIA | N619AA | AA | 39 | 27 | 61.6 | 260 | 15 | 0 | 1.01e+03 | 10 | 2013-01-01 05:00:00 | |
| 2013 | 1 | 1 | 5 | JFK | BQN | N804JB | B6 | 39 | 27 | 61.6 | 260 | 15 | 0 | 1.01e+03 | 10 | 2013-01-01 05:00:00 | |
| 2013 | 1 | 1 | 6 | LGA | ATL | N668DN | DL | 39.9 | 25 | 54.8 | 260 | 16.1 | 23 | 0 | 1.01e+03 | 10 | 2013-01-01 06:00:00 |
| 2013 | 1 | 1 | 5 | EWR | ORD | N39463 | UA | 39 | 28 | 64.4 | 260 | 12.7 | 0 | 1.01e+03 | 10 | 2013-01-01 05:00:00 |
Filter joins
Instead match variables like in regular joints this kind of joints match observations
-
semi_join(x, y) keeps all observations in x that have a match in y.
-
anti_join(x, y) drops all observations in x that have a match in y.
top_dest <- flights %>%
count(dest, sort = TRUE) %>% head(10)
#select top 10 destines
flights %>%
semi_join(top_dest) %>%
head()
#take from flights just the colums that belong to top_dest
| year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2013 | 1 | 1 | 542 | 540 | 2 | 923 | 850 | 33 | AA | 1141 | N619AA | JFK | MIA | 160 | 1.09e+03 | 5 | 40 | 2013-01-01 05:00:00 |
| 2013 | 1 | 1 | 554 | 600 | -6 | 812 | 837 | -25 | DL | 461 | N668DN | LGA | ATL | 116 | 762 | 6 | 0 | 2013-01-01 06:00:00 |
| 2013 | 1 | 1 | 554 | 558 | -4 | 740 | 728 | 12 | UA | 1696 | N39463 | EWR | ORD | 150 | 719 | 5 | 58 | 2013-01-01 05:00:00 |
| 2013 | 1 | 1 | 555 | 600 | -5 | 913 | 854 | 19 | B6 | 507 | N516JB | EWR | FLL | 158 | 1.06e+03 | 6 | 0 | 2013-01-01 06:00:00 |
| 2013 | 1 | 1 | 557 | 600 | -3 | 838 | 846 | -8 | B6 | 79 | N593JB | JFK | MCO | 140 | 944 | 6 | 0 | 2013-01-01 06:00:00 |
| 2013 | 1 | 1 | 558 | 600 | -2 | 753 | 745 | 8 | AA | 301 | N3ALAA | LGA | ORD | 138 | 733 | 6 | 0 | 2013-01-01 06:00:00 |
If you want more information visit R for data science, probably, It’s the best book to learn .